Monte Carlo for a Lennard Jones Fluid - Part 2
Overview
Teaching: 100 min
Exercises: 20 minQuestions
How do I write a Monte Carlo simulation?
Objectives
Use the Metropolis algorithm to sample configurations of LJ particles
Lesson Contents
We will continue today’s script in the notebook from yesterday. Make sure you have the functions
calculate_LJ- written in classcalculate_distance- should be the version written for homework which accounts for periodic boundaries.calculate_total_energy- should be version written for homework which has cutoffread_xyz- provided in classcalculate_tail_correctiion- written for homework
Recall that our objective is to write a Monte Carlo simulation which we can use to estimate properties of a chemical system. Yesterday, we used MC integration to evaluate the area of the unit circle.
We learned that thermodynamic properties can be thought of as very high dimensional integrals.
\[\left<Q\right> = \int_V Q\left(\textbf{r}^N\right) \rho\left(\textbf{r}^N\right) d\textbf{r}^N \label{eq.statMechAverage}\]where \(Q\left(\textbf{r}^N\right)\) is the thermodynamic quantity of interest that depends on only on the configuration, \(\rho\left(\textbf{r}^N\right)\) is the probability density, and \(V\) defines the volume of configuration space over which \(\rho\) has support.
Notice here that unlike our integral yesterday, we have a probability density present in the integral. This depends on the conditions of your system (we will use constant volume, constant temperature). This probability density usually depends on temperature.
In our case, the thermodynamic quantity of interest, \(Q\), was the system potential energy. We used the Lennard Jones equation to build a model of a thermodynamic property (potential energy) of a system of particles.
It is now our task to generate sample configurations of the system. As a consequence, we will be able to measure structural properties of the atoms.
Writing the Monte Carlo Loop
Just like with our first Monte Carlo examples, we will be relying on random numbers to sample our configuration space. This translates into generating possible coordinates for our system. However, due to because our high dimensional integral is dominated by a relatively small region of configurational space, it would be very inefficient just pick random numbers for all of our particles. To overcome this problem, we will use the ideas of importance sampling.
Importance Sampling
A solution to this problem is to sample positions from the desired equilibrium probability density. This will generate relevant configurations more frequently than configurations that have low probability. This idea is known as importance sampling.
A uniform probability density would means that all values are equally likely. Since our system has so many possible system configurations and many of them are not likely to occur, we do not want to waste time/computational resources on those configurations. We will sample positions from the desired equilibrium probability density \(\rho\left(\textbf{r}^N\right)\).
Acceptance Criteria
Now that we know our goal, how will we generate atomic positions which are distributed according to \(\rho\left(\textbf{r}^N\right)\).
We are gong to use the Metropolis-Hasting algorithm for accepting configurations. The Metropolis-Hasting acceptance criteria was first proposed in 1953 by Nicholas Metropolis, and expanded in 1970 by W.K. Hasting. For our system, the probability that we will accept a change of coordinates is expressed with the following equation:
\[P_{acc}(m \rightarrow n) = \text{min} \left[ 1,e^{-\beta \Delta U} \right]\]Here, \(\beta\) is equal to \(1/ T\) (\(T\) is temperature).
Practically, the way this works is that we will start with atoms in some configuration. We calculate the potential energy of that configuration. Then, we make a change in coordinates. If the system is at lower energy, we accept this move as a new configuration. If the system is at a higher energy, we accept or reject that configuration based on comparing \(e^{-\beta \Delta U}\) and a random number.
This is a Markov Chain Monte Carlo method, meaning that acceptance of a random configuration depends on the system’s previous configuration. This is in contrast to our calculation of \(\pi\) where all points were independent of one another.
Flow of Calculations
The workflow for a implementing an MC simulation is shown in the image and summarized below
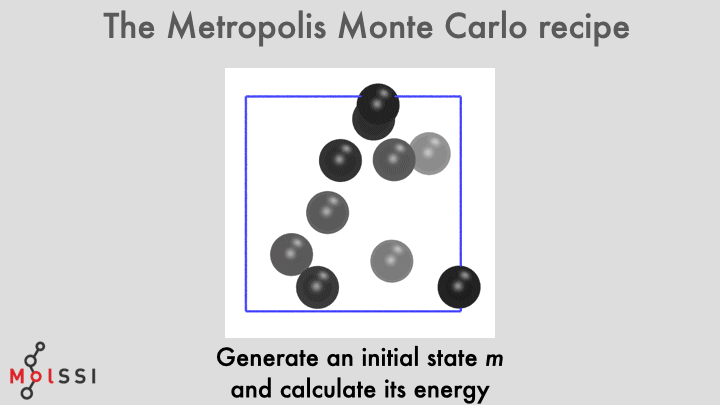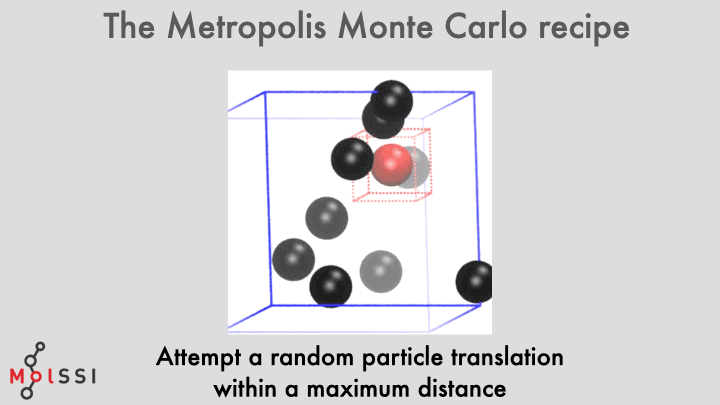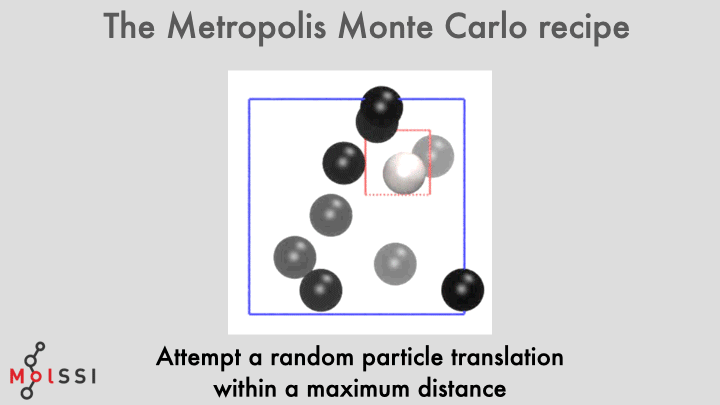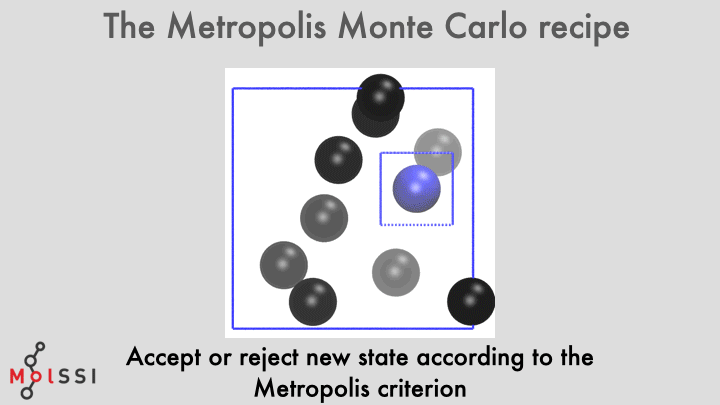
- Generate an initial system state \(m\).
- Choose an atom with uniform probability from \(\{1, \ldots, N\}\) from old state \(m\).
- Propose a new state \(n\) by translating a LJ particle by a uniform random displacement \(\Delta r \sim U(-\Delta x, +\Delta x)\) in each dimension.
- The displacement scale \(\Delta x\) should not be too large as this would likely result in particle overlaps, but should not be too small as this would result in a slow sampling of configurational space.
- The difference in energy between the new and old states is computed.
- The new state is accepted or rejected using the Metropolis criterion.Practically, this can be implemented as follows.
- If a move from \(m\) to \(n\) is “downhill”, \(\beta \Delta U \leq 0\), the move is always accepted. For “uphill” moves, a random number \(\zeta\) is generated uniformly on (0,1).
- If \(\zeta < \exp[-\beta {\Delta U}]\), the move is accepted. Otherwise, the move is rejected.
def accept_or_reject(delta_e, beta):
if delta_e <= 0.0:
accept = True
else:
random_number = random.random()
p_acc = math.exp(-beta*delta_e)
if random_number < p_acc:
accept = True
else:
accept = False
return accept
Sanity check
Let’s do some of our normal sanity checks. We can think of a few things that should be true for this function:
Trueshould be returned for a negativedelta_eTrueshould be returned fordelta_e == 0
For simplicity, we’ll use a beta value of 1 for these two tests:
delta_energy = -1
beta = 1
assert accept_or_reject(delta_energy, beta) is True
delta_energy = 0
beta = 1
assert accept_or_reject(delta_energy, beta) is True
Random Numbers
For delta_e> 0 the function relies on a random number generator. This might seem like it would make the function impossible to test since we never know what numbers we will get. However, we can cause our random function to generate the same set of values every time by using something called a random number seed. A random seed is the starting point in generating random numbers.
Let’s try setting our random seed to 0:
random.seed(0)
Next, generate a random number:
random.random()
0.8444218515250481
Everyone should see the same output for this. Rerun your random number generator again, and you should see 0.7579544029403025.
Set your random number seed to a different number:
random.seed(5)
random.random()
0.6229016948897019
Now, reset your seed to 0:
random.seed(0)
random.random()
0.8444218515250481
You will notice that this is the same random number you got before when you set the seed to 0. Setting a random number seed means that when that seed is set, you will always get the same sequence of random numbers. We will use this for the sanity check of our function.
For writing our test cases, we will use a reduced temperature of 1, or a beta value of 1. We need to set the seed before this test to a value that would cause our criteria to return True. Our acceptance criteria is that the random number we generate has to be less than our probability of acceptance. For \(1/\beta = 1\) and \(\Delta U = 1\), our expression is \(e^{-1}\), or a roughly a 37% chance of the move being accepted. Our random number must be less than this in order for our function to return True.
We already know that when we set the random number seed to zero, our first value will be higher than this. If our function behaves correctly, the function should return False after we set the seed to 0.
random.seed(0)
delta_e = 1
beta = 1
assert accept_or_reject(delta_e, beta) is False
Next, we should find a random seed that returns less than 0.37. Through trial and error, we discover that setting the random seed to be 1 will work:
random.seed(1)
delta_e = 1
beta = 1
assert accept_or_reject(delta_e, beta) is True
After we’ve done these checks, it is a good idea to unset the random seed so that our numbers are random again:
random.seed()
Energy Calculation
In our Monte Carlo loop, we will be calculating the change in energy from a particle movement to decide whether to accept or reject that movement. Previously, we calculated the total pairwise energy for particles. However, when we move a particle, we only need to calculate the energy change for that one particle. We will write a new function for calculating the pairwise interaction energy of a single particle.
def calculate_pair_energy(coordinates, i_particle, box_length, cutoff):
"""
Calculate the interaction energy of a particle with its environment (all other particles in the system)
Parameters
----------
coordinates : list
The coordinates for all particles in the system
i_particle : int
The particle number for which to calculate the energy
cutoff : float
The simulation cutoff. Beyond this distance, interactions are not calculated.
Returns
-------
e_total : float
The pairwise interaction energy of he i_th particle with all other particles in the system.
"""
e_total = 0.0
i_position = coordinates[i_particle]
num_atoms = len(coordinates)
for j_particle in range(num_atoms):
if i_particle != j_particle:
j_position = coordinates[j_particle]
rij = calculate_distance(i_position, j_position, box_length)
if rij < cutoff:
e_pair = calculate_LJ(rij)
e_total += e_pair
return e_total
Sanity Checks
We’ll use the same system of three particles to check this function.
coordinates = [[0, 0, 0], [0, 0, 2**(1/6)], [0, 0, 2*(2**(1/6))]]
assert calculate_pair_energy(coordinates, 1, 10, 3) == -2
assert calculate_pair_energy(coordinates, 0, 10, 3) == calculate_pair_energy(coordinates, 2, 10, 3)
assert calculate_pair_energy(coordinates, 0, 10, 2) == -1
Simulation
Now that we have written the functions we need, we will now write the simulation loop.
We start by defining the simulation parameters. We set the reduced temperature and the number of Monte Carlo steps are going to attempt, as well as the maximum distance we will displace a particle during a Monte Carlo move. We also set a variable here called freq which is the frequency we want to print out energy values.
# Parameters
reduced_temperature = 0.9
num_steps = 5000
max_displacement = 0.1
cutoff = 3.0
freq = 1000
# Calculated quantities
beta = 1 / reduced_temperature
Next, we must have a system of particles to simulate. We will use the reference system from NIST:
# Read initial coordinates
coordinates, box_length = read_xyz('sample_config1.xyz')
num_particles = len(coordinates)
Next, we set our change in energy to 0 and calculate the total energy of the system.
delta_energy = 0
total_energy = calculate_total_energy(coordinates, box_length, cutoff)
total_energy += calculate_tail_correction(num_particles, box_length, cutoff)
We have now finished our initial set-up and are ready for our Monte Carlo loop. We will first create our loop and write in comments what we need to do.
for step in range(num_steps):
# 1. Randomly pick one of N particles
# 2. Calculate the interaction energy of the selected particle with the system and store this value.
# 3. Generate random x, y, z displacement
# 4. Modify coordinate of Nth particle by generated displacements
# 5. Calculate interaction energy of moved particle with the system and store this value.
# 6. Calculate if we accept the move
# 7. if accept, move particle
# 8. print energy if step is multiple of freq
-
Randomly pick one of N particles. For #1 we have to randomly pick one of N particles. This means we need a number on the range of 0 to 800. Take a look at the documentation for the
randommodule and see if you can find a function for this.The function we will use is
rand.randrange. If we addnum_particlesas an argument, it will pick a number between 0 andnum_particles(For our NIST file, this would be between 0 and 799). -
Calculate the interaction energy of the selected particle with the system and store this value.
We will use our
calculate_pair_energyfunction with the random particle index for this. -
Generate a random x, y, z displacement. For #2, we need to generate three random numbers between -max_displacement and max_displacement. By examining the random documentation again, we will find that the function we want to use is
random.uniform. -
Modify coordinate of Nth particle by generated displacements.
We will need to change the position of the particle selected in #1 by the values generated in #2.
-
Calculate interaction energy of moved particle with the system and store this value.
We will use our
calculate_pair_energyfunction with our updated coordinates and the random particle index for this. -
Calculate if we accept the move
Subtract the original system energy from the new system energy, and use the
accept_or_rejectfunction. -
If accept is
True, keep the coordinates, otherwise, rollback to our previous value. -
Print energy if step is multiple of freq. We will use the modulus (
%) operator for this. This operator returns the remainder from division.
We’ll also add an energies list which we’ll append to every time we print the energy. With these considerations, our final MC loop looks like this.
# Parameters
reduced_temperature = 0.9
num_steps = 5000
max_displacement = 0.1
cutoff = 3.0
# Calculated quantities
beta = 1 / reduced_temperature
# Read initial coordinates
coordinates, box_length = read_xyz('sample_config1.xyz')
num_particles = len(coordinates)
delta_energy = 0
total_energy = calculate_total_energy(coordinates, box_length, cutoff)
total_energy += calculate_tail_correction(num_particles, box_length, cutoff)
freq = 1000
steps = []
energies = []
for step in range(num_steps):
# Randomly pick one of N particles
random_particle = random.randrange(num_particles)
# Calculate interaction energy of selected particle with the system
current_energy = calculate_pair_energy(coordinates, random_particle, box_length, cutoff)
# Generate random x, y, z displacement
x_rand = random.uniform(-max_displacement, max_displacement)
y_rand = random.uniform(-max_displacement, max_displacement)
z_rand = random.uniform(-max_displacement, max_displacement)
# Modify coordinate of Nth particle by generated displacements
coordinates[random_particle][0] += x_rand
coordinates[random_particle][1] += y_rand
coordinates[random_particle][2] += z_rand
proposed_energy = calculate_pair_energy(coordinates, random_particle, box_length, cutoff)
# Calculate change in potential energy for particle move.
delta_energy = proposed_energy - current_energy
accept = accept_or_reject(delta_energy, beta)
if accept:
total_energy += delta_energy
n_accept += 1
else:
# Move is not accepted, roll back coordinates
coordinates[random_particle][0] -= x_rand
coordinates[random_particle][1] -= y_rand
coordinates[random_particle][2] -= z_rand
if step % freq == 0:
print(step, total_energy/num_particles)
steps.append(step)
energies.append(total_energy/num_particles)
At the end of your MC run, you will have a list of steps, energies, and your coordinates will be updated to the final MC step.
Key Points
Thermodynamic properties of a system can be thought of as a very high dimensional integral.